1.9. Tabular Data with Pandas#
1.9.1. Introduction#
Pandas is a an open source library providing high-performance, easy-to-use data structures and data analysis tools. Pandas is particularly suited to the analysis of tabular data, i.e. data that can can go into a table. In other words, if you can imagine the data in an Excel spreadsheet, then Pandas is the tool for the job.
A 2017 recent analysis of questions from Stack Overflow showed that python was the fastest growing and most widely used programming language in the world (in developed countries). As of 2021, the growth has now leveled off, but Python remains at the top.

Link to generate your own version of this figure
A follow-up analysis showed that this growth is driven by the data science packages such as numpy, matplotlib, and especially pandas.
The exponential growth of pandas is due to the fact that it just works. It saves you time and helps you do science more efficiently and effectively.
Pandas capabilities (from the Pandas website):
A fast and efficient DataFrame object for data manipulation with integrated indexing;
Tools for reading and writing data between in-memory data structures and different formats: CSV and text files, Microsoft Excel, SQL databases, and the fast HDF5 format;
Intelligent data alignment and integrated handling of missing data: gain automatic label-based alignment in computations and easily manipulate messy data into an orderly form;
Flexible reshaping and pivoting of data sets;
Intelligent label-based slicing, fancy indexing, and subsetting of large data sets;
Columns can be inserted and deleted from data structures for size mutability;
Aggregating or transforming data with a powerful group by engine allowing split-apply-combine operations on data sets;
High performance merging and joining of data sets;
Hierarchical axis indexing provides an intuitive way of working with high-dimensional data in a lower-dimensional data structure;
Time series-functionality: date range generation and frequency conversion, moving window statistics, moving window linear regressions, date shifting and lagging. Even create domain-specific time offsets and join time series without losing data;
Highly optimized for performance, with critical code paths written in Cython or C.
In this notebook, we will go over the basic capabilities of Pandas. It is a very deep library, and you will need to dig into the documentation for more advanced usage.
Pandas was created by Wes McKinney. Many of the examples here are drawn from Wes McKinney’s book Python for Data Analysis, which includes “a GitHub repo of code samples.
1.9.2. Pandas Data Structures: Series#
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
%matplotlib inline
A Series represents a one-dimensional array of data. The main difference between a Series and numpy array is that a Series has an index. The index contains the labels that we use to access the data.
There are many ways to create a Series. We will just show a few.
(Data are from the NASA Planetary Fact Sheet.)
names = ['Mercury', 'Venus', 'Earth']
values = [0.3e24, 4.87e24, 5.97e24]
masses = pd.Series(values, index=names)
masses
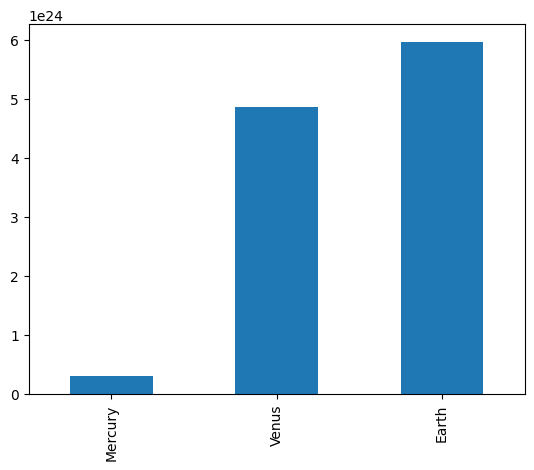
Mercury 3.000000e+23
Venus 4.870000e+24
Earth 5.970000e+24
dtype: float64
# Series have built in plotting methods.
masses.plot(kind='bar')
<Axes: >
# Arithmetic operations and most `numpy` function can be applied to Series.
# An important point is that the Series keep their index during such operations.
np.log(masses) / masses**2
Mercury 6.006452e-46
Venus 2.396820e-48
Earth 1.600655e-48
dtype: float64
# We can access the underlying index object if we need to:
masses.index
Index(['Mercury', 'Venus', 'Earth'], dtype='object')
1.9.2.1. Indexing#
# We can get values back out using the index via the `.loc` attribute
masses.loc['Earth']
5.97e+24
# Or by raw position using `.iloc`
masses.iloc[2]
5.97e+24
# We can pass a list or array to loc to get multiple rows back:
masses.loc[['Venus', 'Earth']]
Venus 4.870000e+24
Earth 5.970000e+24
dtype: float64
# And we can even use slice notation
masses.loc['Mercury':'Earth']
Mercury 3.000000e+23
Venus 4.870000e+24
Earth 5.970000e+24
dtype: float64
masses.iloc[:2]
Mercury 3.000000e+23
Venus 4.870000e+24
dtype: float64
# If we need to, we can always get the raw data back out as well
masses.values # a numpy array
array([3.00e+23, 4.87e+24, 5.97e+24])
masses.index # a pandas Index object
Index(['Mercury', 'Venus', 'Earth'], dtype='object')
1.9.3. Pandas Data Structures: DataFrame#
There is a lot more to Series, but they are limit to a single “column”. A more useful Pandas data structure is the DataFrame. A DataFrame is basically a bunch of series that share the same index. It’s a lot like a table in a spreadsheet.
Below we create a DataFrame.
# first we create a dictionary
data = {'mass': [0.3e24, 4.87e24, 5.97e24], # kg
'diameter': [4879e3, 12_104e3, 12_756e3], # m
'rotation_period': [1407.6, np.nan, 23.9] # h
}
df = pd.DataFrame(data, index=['Mercury', 'Venus', 'Earth'])
df
| mass | diameter | rotation_period | |
|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 |
| Venus | 4.870000e+24 | 12104000.0 | NaN |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 |
Pandas handles missing data very elegantly, keeping track of it through all calculations.
df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 3 entries, Mercury to Earth
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mass 3 non-null float64
1 diameter 3 non-null float64
2 rotation_period 2 non-null float64
dtypes: float64(3)
memory usage: 96.0+ bytes
A wide range of statistical functions are available on both Series and DataFrames.
print(df.min())
print(df.mean())
print(df.std())
print(df.describe())
mass 3.000000e+23
diameter 4.879000e+06
rotation_period 2.390000e+01
dtype: float64
mass 3.713333e+24
diameter 9.913000e+06
rotation_period 7.157500e+02
dtype: float64
mass 3.006765e+24
diameter 4.371744e+06
rotation_period 9.784237e+02
dtype: float64
mass diameter rotation_period
count 3.000000e+00 3.000000e+00 2.000000
mean 3.713333e+24 9.913000e+06 715.750000
std 3.006765e+24 4.371744e+06 978.423653
min 3.000000e+23 4.879000e+06 23.900000
25% 2.585000e+24 8.491500e+06 369.825000
50% 4.870000e+24 1.210400e+07 715.750000
75% 5.420000e+24 1.243000e+07 1061.675000
max 5.970000e+24 1.275600e+07 1407.600000
# We can get a single column as a `Series` using python’s `getitem` syntax on the `DataFrame` object.
print(df['mass'])
# …or using attribute syntax.
print(df.mass)
Mercury 3.000000e+23
Venus 4.870000e+24
Earth 5.970000e+24
Name: mass, dtype: float64
Mercury 3.000000e+23
Venus 4.870000e+24
Earth 5.970000e+24
Name: mass, dtype: float64
# Indexing works very similar to series
print(df.loc['Earth'])
print(df.iloc[2])
mass 5.970000e+24
diameter 1.275600e+07
rotation_period 2.390000e+01
Name: Earth, dtype: float64
mass 5.970000e+24
diameter 1.275600e+07
rotation_period 2.390000e+01
Name: Earth, dtype: float64
# But we can also specify the column we want to access
df.loc['Earth', 'mass']
5.97e+24
df.iloc[0:2, 0]
Mercury 3.000000e+23
Venus 4.870000e+24
Name: mass, dtype: float64
If we make a calculation using columns from the DataFrame, it will keep the same index:
volume = 4/3 * np.pi * (df.diameter/2)**3
df.mass / volume
Mercury 4933.216530
Venus 5244.977070
Earth 5493.285577
dtype: float64
Which we can easily add as another column to the DataFrame:
df['density'] = df.mass / volume
df
| mass | diameter | rotation_period | density | |
|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 |
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 |
1.9.4. Merging Data#
Pandas supports a wide range of methods for merging different datasets. These are described extensively in the documentation. Here we just give a few examples.
temperature = pd.Series([167, 464, 15, -65],
index=['Mercury', 'Venus', 'Earth', 'Mars'],
name='temperature')
temperature
Mercury 167
Venus 464
Earth 15
Mars -65
Name: temperature, dtype: int64
# returns a new DataFrame
df.join(temperature)
| mass | diameter | rotation_period | density | temperature | |
|---|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 | 167 |
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 | 464 |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 | 15 |
# returns a new DataFrame
df.join(temperature, how='right')
| mass | diameter | rotation_period | density | temperature | |
|---|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 | 167 |
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 | 464 |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 | 15 |
| Mars | NaN | NaN | NaN | NaN | -65 |
# returns a new DataFrame
everyone = df.reindex(['Mercury', 'Venus', 'Earth', 'Mars'])
everyone
| mass | diameter | rotation_period | density | |
|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 |
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 |
| Mars | NaN | NaN | NaN | NaN |
We can also index using a boolean series. This is very useful:
adults = df[df.mass > 4e24]
adults
| mass | diameter | rotation_period | density | |
|---|---|---|---|---|
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 |
df['is_big'] = df.mass > 4e24
df
| mass | diameter | rotation_period | density | is_big | |
|---|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 | False |
| Venus | 4.870000e+24 | 12104000.0 | NaN | 5244.977070 | True |
| Earth | 5.970000e+24 | 12756000.0 | 23.9 | 5493.285577 | True |
1.9.5. Modifying Values#
We often want to modify values in a dataframe based on some rule. To modify values, we need to use .loc or .iloc
df.loc['Earth', 'mass'] = 5.98+24
df.loc['Venus', 'diameter'] += 1
df
| mass | diameter | rotation_period | density | is_big | |
|---|---|---|---|---|---|
| Mercury | 3.000000e+23 | 4879000.0 | 1407.6 | 4933.216530 | False |
| Venus | 4.870000e+24 | 12104001.0 | NaN | 5244.977070 | True |
| Earth | 2.998000e+01 | 12756000.0 | 23.9 | 5493.285577 | True |
1.9.6. Plotting#
DataFrames have all kinds of useful plotting built in.
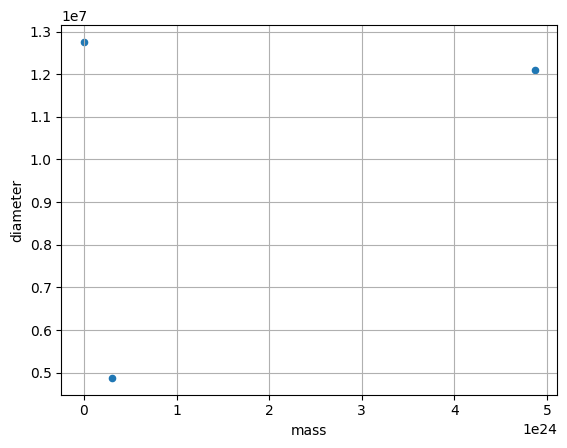
df.plot(kind='scatter', x='mass', y='diameter', grid=True)
<Axes: xlabel='mass', ylabel='diameter'>
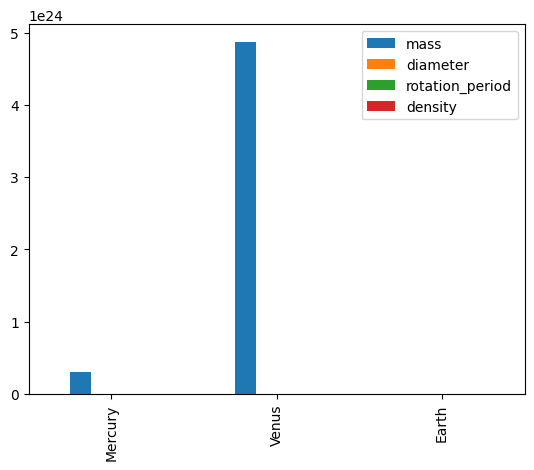
df.plot(kind='bar')
<Axes: >
1.9.7. Time Indexes#
Indexes are very powerful. They are a big part of why Pandas is so useful. There are different indexes for different types of data. Time indexes are especially great!
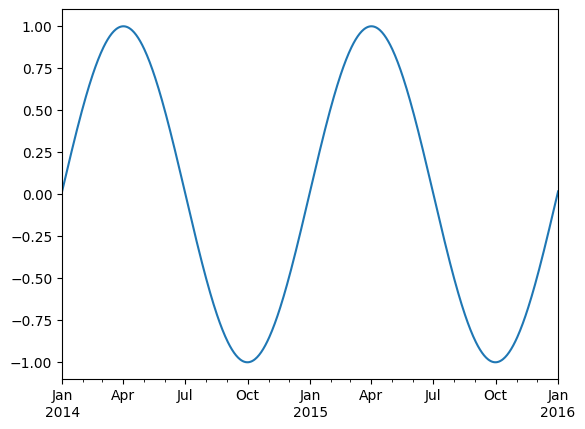
two_years = pd.date_range(start='2014-01-01', end='2016-01-01', freq='D')
timeseries = pd.Series(np.sin(2 *np.pi *two_years.dayofyear / 365),
index=two_years)
timeseries.plot()
<Axes: >
We can use python’s slicing notation inside .loc to select a date range.
timeseries.loc['2015-01-01':'2015-07-01'].plot()
<Axes: >
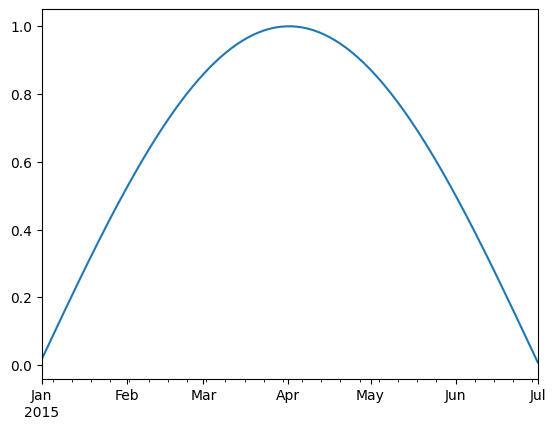
timeseries.index.month
Index([ 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
...
12, 12, 12, 12, 12, 12, 12, 12, 12, 1],
dtype='int32', length=731)
timeseries.index.day
Index([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
...
23, 24, 25, 26, 27, 28, 29, 30, 31, 1],
dtype='int32', length=731)
1.9.8. Reading Data Files: Weather Station Data#
In this example, we will use NOAA weather station data from https://www.ncei.noaa.gov/products/land-based-station.
The details of files we are going to read are described in this README file.
import pooch
POOCH = pooch.create(
path=pooch.os_cache("noaa-data"),
base_url="doi:10.5281/zenodo.5564850/",
registry={
"data.txt": "md5:5129dcfd19300eb8d4d8d1673fcfbcb4",
},
)
datafile = POOCH.fetch("data.txt")
datafile
'C:\\Users\\tbeucler\\AppData\\Local\\noaa-data\\noaa-data\\Cache\\data.txt'
! head '/root/.cache/noaa-data/data.txt' # Replace this value with the download path indicated above
'head' is not recognized as an internal or external command,
operable program or batch file.
We now have a text file on our hard drive called data.txt.
To read it into pandas, we will use the read_csv function. This function is incredibly complex and powerful. You can use it to extract data from almost any text file. However, you need to understand how to use its various options.
With no options, this is what we get.
df = pd.read_csv(datafile)
df.head()
| WBANNO LST_DATE CRX_VN LONGITUDE LATITUDE T_DAILY_MAX T_DAILY_MIN T_DAILY_MEAN T_DAILY_AVG P_DAILY_CALC SOLARAD_DAILY SUR_TEMP_DAILY_TYPE SUR_TEMP_DAILY_MAX SUR_TEMP_DAILY_MIN SUR_TEMP_DAILY_AVG RH_DAILY_MAX RH_DAILY_MIN RH_DAILY_AVG SOIL_MOISTURE_5_DAILY SOIL_MOISTURE_10_DAILY SOIL_MOISTURE_20_DAILY SOIL_MOISTURE_50_DAILY SOIL_MOISTURE_100_DAILY SOIL_TEMP_5_DAILY SOIL_TEMP_10_DAILY SOIL_TEMP_20_DAILY SOIL_TEMP_50_DAILY SOIL_TEMP_100_DAILY | |
|---|---|
| 0 | 64756 20170101 2.422 -73.74 41.79 6.6 ... |
| 1 | 64756 20170102 2.422 -73.74 41.79 4.0 ... |
| 2 | 64756 20170103 2.422 -73.74 41.79 4.9 ... |
| 3 | 64756 20170104 2.422 -73.74 41.79 8.7 ... |
| 4 | 64756 20170105 2.422 -73.74 41.79 -0.5 ... |
Pandas failed to identify the different columns. This is because it was expecting standard CSV (comma-separated values) file. In our file, instead, the values are separated by whitespace. And not a single whilespace–the amount of whitespace between values varies. We can tell pandas this using the sep keyword.
df = pd.read_csv(datafile, sep='\s+')
df.head()
| WBANNO | LST_DATE | CRX_VN | LONGITUDE | LATITUDE | T_DAILY_MAX | T_DAILY_MIN | T_DAILY_MEAN | T_DAILY_AVG | P_DAILY_CALC | ... | SOIL_MOISTURE_5_DAILY | SOIL_MOISTURE_10_DAILY | SOIL_MOISTURE_20_DAILY | SOIL_MOISTURE_50_DAILY | SOIL_MOISTURE_100_DAILY | SOIL_TEMP_5_DAILY | SOIL_TEMP_10_DAILY | SOIL_TEMP_20_DAILY | SOIL_TEMP_50_DAILY | SOIL_TEMP_100_DAILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 64756 | 20170101 | 2.422 | -73.74 | 41.79 | 6.6 | -5.4 | 0.6 | 2.2 | 0.0 | ... | -99.0 | -99.0 | 0.207 | 0.152 | 0.175 | -0.1 | 0.0 | 0.6 | 1.5 | 3.4 |
| 1 | 64756 | 20170102 | 2.422 | -73.74 | 41.79 | 4.0 | -6.8 | -1.4 | -1.2 | 0.0 | ... | -99.0 | -99.0 | 0.205 | 0.151 | 0.173 | -0.2 | 0.0 | 0.6 | 1.5 | 3.3 |
| 2 | 64756 | 20170103 | 2.422 | -73.74 | 41.79 | 4.9 | 0.7 | 2.8 | 2.7 | 13.1 | ... | -99.0 | -99.0 | 0.205 | 0.150 | 0.173 | -0.1 | 0.0 | 0.5 | 1.5 | 3.3 |
| 3 | 64756 | 20170104 | 2.422 | -73.74 | 41.79 | 8.7 | -1.6 | 3.6 | 3.5 | 1.3 | ... | -99.0 | -99.0 | 0.215 | 0.153 | 0.174 | -0.1 | 0.0 | 0.5 | 1.5 | 3.2 |
| 4 | 64756 | 20170105 | 2.422 | -73.74 | 41.79 | -0.5 | -4.6 | -2.5 | -2.8 | 0.0 | ... | -99.0 | -99.0 | 0.215 | 0.154 | 0.177 | -0.1 | 0.0 | 0.5 | 1.4 | 3.1 |
5 rows × 28 columns
Great! It worked.
If we look closely, we will see there are lots of -99 and -9999 values in the file. The README file tells us that these are values used to represent missing data. Let’s tell this to pandas.
df = pd.read_csv(datafile, sep='\s+', na_values=[-9999.0, -99.0])
df.head()
| WBANNO | LST_DATE | CRX_VN | LONGITUDE | LATITUDE | T_DAILY_MAX | T_DAILY_MIN | T_DAILY_MEAN | T_DAILY_AVG | P_DAILY_CALC | ... | SOIL_MOISTURE_5_DAILY | SOIL_MOISTURE_10_DAILY | SOIL_MOISTURE_20_DAILY | SOIL_MOISTURE_50_DAILY | SOIL_MOISTURE_100_DAILY | SOIL_TEMP_5_DAILY | SOIL_TEMP_10_DAILY | SOIL_TEMP_20_DAILY | SOIL_TEMP_50_DAILY | SOIL_TEMP_100_DAILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 64756 | 20170101 | 2.422 | -73.74 | 41.79 | 6.6 | -5.4 | 0.6 | 2.2 | 0.0 | ... | NaN | NaN | 0.207 | 0.152 | 0.175 | -0.1 | 0.0 | 0.6 | 1.5 | 3.4 |
| 1 | 64756 | 20170102 | 2.422 | -73.74 | 41.79 | 4.0 | -6.8 | -1.4 | -1.2 | 0.0 | ... | NaN | NaN | 0.205 | 0.151 | 0.173 | -0.2 | 0.0 | 0.6 | 1.5 | 3.3 |
| 2 | 64756 | 20170103 | 2.422 | -73.74 | 41.79 | 4.9 | 0.7 | 2.8 | 2.7 | 13.1 | ... | NaN | NaN | 0.205 | 0.150 | 0.173 | -0.1 | 0.0 | 0.5 | 1.5 | 3.3 |
| 3 | 64756 | 20170104 | 2.422 | -73.74 | 41.79 | 8.7 | -1.6 | 3.6 | 3.5 | 1.3 | ... | NaN | NaN | 0.215 | 0.153 | 0.174 | -0.1 | 0.0 | 0.5 | 1.5 | 3.2 |
| 4 | 64756 | 20170105 | 2.422 | -73.74 | 41.79 | -0.5 | -4.6 | -2.5 | -2.8 | 0.0 | ... | NaN | NaN | 0.215 | 0.154 | 0.177 | -0.1 | 0.0 | 0.5 | 1.4 | 3.1 |
5 rows × 28 columns
Wonderful. The missing data is now represented by NaN.
What data types did pandas infer?
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 365 entries, 0 to 364
Data columns (total 28 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 WBANNO 365 non-null int64
1 LST_DATE 365 non-null int64
2 CRX_VN 365 non-null float64
3 LONGITUDE 365 non-null float64
4 LATITUDE 365 non-null float64
5 T_DAILY_MAX 364 non-null float64
6 T_DAILY_MIN 364 non-null float64
7 T_DAILY_MEAN 364 non-null float64
8 T_DAILY_AVG 364 non-null float64
9 P_DAILY_CALC 364 non-null float64
10 SOLARAD_DAILY 364 non-null float64
11 SUR_TEMP_DAILY_TYPE 365 non-null object
12 SUR_TEMP_DAILY_MAX 364 non-null float64
13 SUR_TEMP_DAILY_MIN 364 non-null float64
14 SUR_TEMP_DAILY_AVG 364 non-null float64
15 RH_DAILY_MAX 364 non-null float64
16 RH_DAILY_MIN 364 non-null float64
17 RH_DAILY_AVG 364 non-null float64
18 SOIL_MOISTURE_5_DAILY 317 non-null float64
19 SOIL_MOISTURE_10_DAILY 317 non-null float64
20 SOIL_MOISTURE_20_DAILY 336 non-null float64
21 SOIL_MOISTURE_50_DAILY 364 non-null float64
22 SOIL_MOISTURE_100_DAILY 359 non-null float64
23 SOIL_TEMP_5_DAILY 364 non-null float64
24 SOIL_TEMP_10_DAILY 364 non-null float64
25 SOIL_TEMP_20_DAILY 364 non-null float64
26 SOIL_TEMP_50_DAILY 364 non-null float64
27 SOIL_TEMP_100_DAILY 364 non-null float64
dtypes: float64(25), int64(2), object(1)
memory usage: 80.0+ KB
One problem here is that pandas did not recognize the LST_DATE column as a date. Let’s help it.
df = pd.read_csv(datafile, sep='\s+',
na_values=[-9999.0, -99.0],
parse_dates=[1])
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 365 entries, 0 to 364
Data columns (total 28 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 WBANNO 365 non-null int64
1 LST_DATE 365 non-null datetime64[ns]
2 CRX_VN 365 non-null float64
3 LONGITUDE 365 non-null float64
4 LATITUDE 365 non-null float64
5 T_DAILY_MAX 364 non-null float64
6 T_DAILY_MIN 364 non-null float64
7 T_DAILY_MEAN 364 non-null float64
8 T_DAILY_AVG 364 non-null float64
9 P_DAILY_CALC 364 non-null float64
10 SOLARAD_DAILY 364 non-null float64
11 SUR_TEMP_DAILY_TYPE 365 non-null object
12 SUR_TEMP_DAILY_MAX 364 non-null float64
13 SUR_TEMP_DAILY_MIN 364 non-null float64
14 SUR_TEMP_DAILY_AVG 364 non-null float64
15 RH_DAILY_MAX 364 non-null float64
16 RH_DAILY_MIN 364 non-null float64
17 RH_DAILY_AVG 364 non-null float64
18 SOIL_MOISTURE_5_DAILY 317 non-null float64
19 SOIL_MOISTURE_10_DAILY 317 non-null float64
20 SOIL_MOISTURE_20_DAILY 336 non-null float64
21 SOIL_MOISTURE_50_DAILY 364 non-null float64
22 SOIL_MOISTURE_100_DAILY 359 non-null float64
23 SOIL_TEMP_5_DAILY 364 non-null float64
24 SOIL_TEMP_10_DAILY 364 non-null float64
25 SOIL_TEMP_20_DAILY 364 non-null float64
26 SOIL_TEMP_50_DAILY 364 non-null float64
27 SOIL_TEMP_100_DAILY 364 non-null float64
dtypes: datetime64[ns](1), float64(25), int64(1), object(1)
memory usage: 80.0+ KB
# It worked! Finally, let’s tell pandas to use the date column as the index.
df = df.set_index('LST_DATE')
df.head()
| WBANNO | CRX_VN | LONGITUDE | LATITUDE | T_DAILY_MAX | T_DAILY_MIN | T_DAILY_MEAN | T_DAILY_AVG | P_DAILY_CALC | SOLARAD_DAILY | ... | SOIL_MOISTURE_5_DAILY | SOIL_MOISTURE_10_DAILY | SOIL_MOISTURE_20_DAILY | SOIL_MOISTURE_50_DAILY | SOIL_MOISTURE_100_DAILY | SOIL_TEMP_5_DAILY | SOIL_TEMP_10_DAILY | SOIL_TEMP_20_DAILY | SOIL_TEMP_50_DAILY | SOIL_TEMP_100_DAILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LST_DATE | |||||||||||||||||||||
| 2017-01-01 | 64756 | 2.422 | -73.74 | 41.79 | 6.6 | -5.4 | 0.6 | 2.2 | 0.0 | 8.68 | ... | NaN | NaN | 0.207 | 0.152 | 0.175 | -0.1 | 0.0 | 0.6 | 1.5 | 3.4 |
| 2017-01-02 | 64756 | 2.422 | -73.74 | 41.79 | 4.0 | -6.8 | -1.4 | -1.2 | 0.0 | 2.08 | ... | NaN | NaN | 0.205 | 0.151 | 0.173 | -0.2 | 0.0 | 0.6 | 1.5 | 3.3 |
| 2017-01-03 | 64756 | 2.422 | -73.74 | 41.79 | 4.9 | 0.7 | 2.8 | 2.7 | 13.1 | 0.68 | ... | NaN | NaN | 0.205 | 0.150 | 0.173 | -0.1 | 0.0 | 0.5 | 1.5 | 3.3 |
| 2017-01-04 | 64756 | 2.422 | -73.74 | 41.79 | 8.7 | -1.6 | 3.6 | 3.5 | 1.3 | 2.85 | ... | NaN | NaN | 0.215 | 0.153 | 0.174 | -0.1 | 0.0 | 0.5 | 1.5 | 3.2 |
| 2017-01-05 | 64756 | 2.422 | -73.74 | 41.79 | -0.5 | -4.6 | -2.5 | -2.8 | 0.0 | 4.90 | ... | NaN | NaN | 0.215 | 0.154 | 0.177 | -0.1 | 0.0 | 0.5 | 1.4 | 3.1 |
5 rows × 27 columns
df.loc['2017-08-07']
WBANNO 64756
CRX_VN 2.422
LONGITUDE -73.74
LATITUDE 41.79
T_DAILY_MAX 19.3
T_DAILY_MIN 12.3
T_DAILY_MEAN 15.8
T_DAILY_AVG 16.3
P_DAILY_CALC 4.9
SOLARAD_DAILY 3.93
SUR_TEMP_DAILY_TYPE C
SUR_TEMP_DAILY_MAX 22.3
SUR_TEMP_DAILY_MIN 11.9
SUR_TEMP_DAILY_AVG 17.7
RH_DAILY_MAX 94.7
RH_DAILY_MIN 76.4
RH_DAILY_AVG 89.5
SOIL_MOISTURE_5_DAILY 0.148
SOIL_MOISTURE_10_DAILY 0.113
SOIL_MOISTURE_20_DAILY 0.094
SOIL_MOISTURE_50_DAILY 0.114
SOIL_MOISTURE_100_DAILY 0.151
SOIL_TEMP_5_DAILY 21.4
SOIL_TEMP_10_DAILY 21.7
SOIL_TEMP_20_DAILY 22.1
SOIL_TEMP_50_DAILY 22.2
SOIL_TEMP_100_DAILY 21.5
Name: 2017-08-07 00:00:00, dtype: object
df.loc['2017-07-01':'2017-07-11']
| WBANNO | CRX_VN | LONGITUDE | LATITUDE | T_DAILY_MAX | T_DAILY_MIN | T_DAILY_MEAN | T_DAILY_AVG | P_DAILY_CALC | SOLARAD_DAILY | ... | SOIL_MOISTURE_5_DAILY | SOIL_MOISTURE_10_DAILY | SOIL_MOISTURE_20_DAILY | SOIL_MOISTURE_50_DAILY | SOIL_MOISTURE_100_DAILY | SOIL_TEMP_5_DAILY | SOIL_TEMP_10_DAILY | SOIL_TEMP_20_DAILY | SOIL_TEMP_50_DAILY | SOIL_TEMP_100_DAILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LST_DATE | |||||||||||||||||||||
| 2017-07-01 | 64756 | 2.422 | -73.74 | 41.79 | 28.0 | 19.7 | 23.9 | 23.8 | 0.2 | 19.28 | ... | 0.157 | 0.136 | 0.144 | 0.129 | 0.163 | 25.7 | 25.4 | 23.7 | 21.9 | 19.9 |
| 2017-07-02 | 64756 | 2.422 | -73.74 | 41.79 | 29.8 | 18.4 | 24.1 | 23.7 | 4.0 | 27.67 | ... | 0.146 | 0.135 | 0.143 | 0.129 | 0.162 | 26.8 | 26.4 | 24.5 | 22.3 | 20.1 |
| 2017-07-03 | 64756 | 2.422 | -73.74 | 41.79 | 28.3 | 15.0 | 21.7 | 21.4 | 0.0 | 27.08 | ... | 0.141 | 0.132 | 0.139 | 0.128 | 0.162 | 26.4 | 26.3 | 24.8 | 22.8 | 20.3 |
| 2017-07-04 | 64756 | 2.422 | -73.74 | 41.79 | 26.8 | 12.6 | 19.7 | 20.0 | 0.0 | 29.45 | ... | 0.131 | 0.126 | 0.136 | 0.126 | 0.161 | 25.9 | 25.8 | 24.6 | 22.9 | 20.6 |
| 2017-07-05 | 64756 | 2.422 | -73.74 | 41.79 | 28.0 | 11.9 | 20.0 | 20.7 | 0.0 | 26.90 | ... | 0.116 | 0.114 | 0.131 | 0.125 | 0.161 | 25.3 | 25.3 | 24.2 | 22.8 | 20.7 |
| 2017-07-06 | 64756 | 2.422 | -73.74 | 41.79 | 25.7 | 14.3 | 20.0 | 20.3 | 0.0 | 19.03 | ... | 0.105 | 0.104 | 0.126 | 0.124 | 0.160 | 24.7 | 24.7 | 23.9 | 22.7 | 20.9 |
| 2017-07-07 | 64756 | 2.422 | -73.74 | 41.79 | 25.8 | 16.8 | 21.3 | 20.0 | 11.5 | 13.88 | ... | 0.114 | 0.100 | 0.123 | 0.123 | 0.160 | 24.2 | 24.2 | 23.4 | 22.4 | 20.8 |
| 2017-07-08 | 64756 | 2.422 | -73.74 | 41.79 | 29.0 | 15.3 | 22.1 | 21.5 | 0.0 | 21.92 | ... | 0.130 | 0.106 | 0.122 | 0.123 | 0.159 | 25.5 | 25.3 | 23.9 | 22.4 | 20.8 |
| 2017-07-09 | 64756 | 2.422 | -73.74 | 41.79 | 26.3 | 10.9 | 18.6 | 19.4 | 0.0 | 29.72 | ... | 0.119 | 0.103 | 0.119 | 0.121 | 0.158 | 24.8 | 24.8 | 23.8 | 22.5 | 20.8 |
| 2017-07-10 | 64756 | 2.422 | -73.74 | 41.79 | 27.6 | 11.8 | 19.7 | 21.3 | 0.0 | 23.67 | ... | 0.105 | 0.096 | 0.113 | 0.120 | 0.158 | 24.7 | 24.7 | 23.6 | 22.5 | 20.9 |
| 2017-07-11 | 64756 | 2.422 | -73.74 | 41.79 | 27.4 | 19.2 | 23.3 | 22.6 | 8.5 | 17.79 | ... | 0.106 | 0.093 | 0.110 | 0.120 | 0.157 | 25.6 | 25.4 | 24.1 | 22.6 | 20.9 |
11 rows × 27 columns
1.9.9. Quick statistics#
df.describe()
| WBANNO | CRX_VN | LONGITUDE | LATITUDE | T_DAILY_MAX | T_DAILY_MIN | T_DAILY_MEAN | T_DAILY_AVG | P_DAILY_CALC | SOLARAD_DAILY | ... | SOIL_MOISTURE_5_DAILY | SOIL_MOISTURE_10_DAILY | SOIL_MOISTURE_20_DAILY | SOIL_MOISTURE_50_DAILY | SOIL_MOISTURE_100_DAILY | SOIL_TEMP_5_DAILY | SOIL_TEMP_10_DAILY | SOIL_TEMP_20_DAILY | SOIL_TEMP_50_DAILY | SOIL_TEMP_100_DAILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 365.0 | 365.000000 | 365.00 | 3.650000e+02 | 364.000000 | 364.000000 | 364.000000 | 364.000000 | 364.000000 | 364.000000 | ... | 317.000000 | 317.000000 | 336.000000 | 364.000000 | 359.000000 | 364.000000 | 364.000000 | 364.000000 | 364.000000 | 364.000000 |
| mean | 64756.0 | 2.470767 | -73.74 | 4.179000e+01 | 15.720055 | 4.037912 | 9.876374 | 9.990110 | 2.797802 | 13.068187 | ... | 0.189498 | 0.183991 | 0.165470 | 0.140192 | 0.160630 | 12.312637 | 12.320604 | 12.060165 | 11.978022 | 11.915659 |
| std | 0.0 | 0.085997 | 0.00 | 7.115181e-15 | 10.502087 | 9.460676 | 9.727451 | 9.619168 | 7.238628 | 7.953074 | ... | 0.052031 | 0.054113 | 0.043989 | 0.020495 | 0.016011 | 9.390034 | 9.338176 | 8.767752 | 8.078346 | 7.187317 |
| min | 64756.0 | 2.422000 | -73.74 | 4.179000e+01 | -12.300000 | -21.800000 | -17.000000 | -16.700000 | 0.000000 | 0.100000 | ... | 0.075000 | 0.078000 | 0.087000 | 0.101000 | 0.117000 | -0.700000 | -0.400000 | 0.200000 | 0.900000 | 1.900000 |
| 25% | 64756.0 | 2.422000 | -73.74 | 4.179000e+01 | 6.900000 | -2.775000 | 2.100000 | 2.275000 | 0.000000 | 6.225000 | ... | 0.152000 | 0.139000 | 0.118750 | 0.118000 | 0.154000 | 2.225000 | 2.000000 | 2.475000 | 3.300000 | 4.100000 |
| 50% | 64756.0 | 2.422000 | -73.74 | 4.179000e+01 | 17.450000 | 4.350000 | 10.850000 | 11.050000 | 0.000000 | 12.865000 | ... | 0.192000 | 0.198000 | 0.183000 | 0.147500 | 0.165000 | 13.300000 | 13.350000 | 13.100000 | 12.850000 | 11.600000 |
| 75% | 64756.0 | 2.422000 | -73.74 | 4.179000e+01 | 24.850000 | 11.900000 | 18.150000 | 18.450000 | 1.400000 | 19.740000 | ... | 0.234000 | 0.227000 | 0.203000 | 0.157000 | 0.173000 | 21.025000 | 21.125000 | 20.400000 | 19.800000 | 19.325000 |
| max | 64756.0 | 2.622000 | -73.74 | 4.179000e+01 | 33.400000 | 20.700000 | 25.700000 | 26.700000 | 65.700000 | 29.910000 | ... | 0.296000 | 0.321000 | 0.235000 | 0.182000 | 0.192000 | 27.600000 | 27.400000 | 25.600000 | 24.100000 | 22.100000 |
8 rows × 26 columns
1.9.10. Plotting Values#
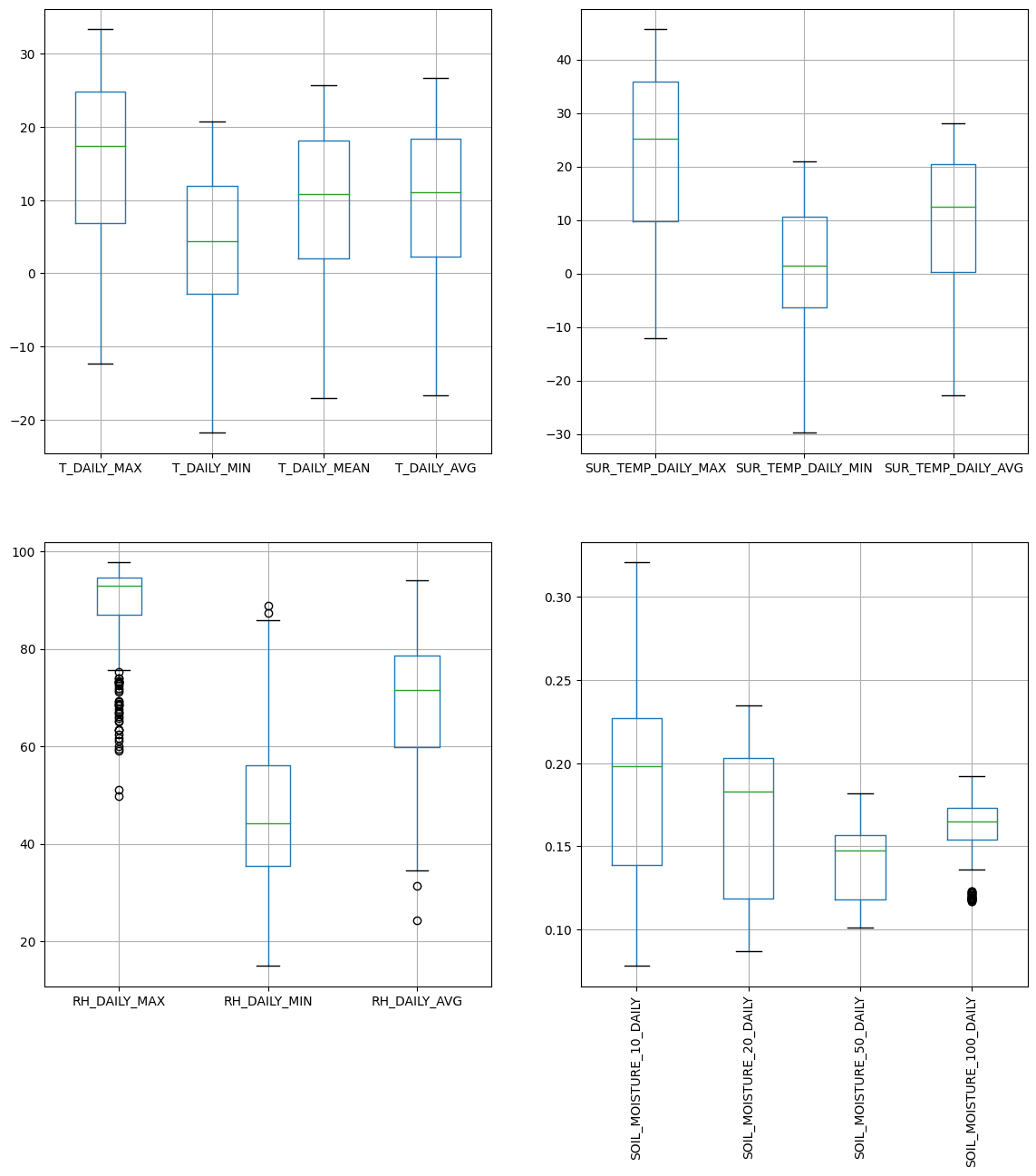
Pandas integrates a convenient boxplot function:
fig, ax = plt.subplots(ncols=2, nrows=2, figsize=(14,14))
df.iloc[:, 4:8].boxplot(ax=ax[0,0])
df.iloc[:, 10:14].boxplot(ax=ax[0,1])
df.iloc[:, 14:17].boxplot(ax=ax[1,0])
df.iloc[:, 18:22].boxplot(ax=ax[1,1])
ax[1, 1].set_xticklabels(ax[1, 1].get_xticklabels(), rotation=90);
# Pandas is "TIME-AWARE"
df.T_DAILY_MEAN.plot()
<Axes: xlabel='LST_DATE'>
Note: we could also manually create an axis and plot into it.
fig, ax = plt.subplots()
df.T_DAILY_MEAN.plot(ax=ax)
ax.set_title('Pandas Made This!')
Text(0.5, 1.0, 'Pandas Made This!')
df[['T_DAILY_MIN', 'T_DAILY_MEAN', 'T_DAILY_MAX']].plot()
<Axes: xlabel='LST_DATE'>
1.9.11. Resampling#
Since pandas understands time, we can use it to do resampling. The frequency string of each date offset is listed in the time series documentation.
# monthly reampler object
rs_obj = df.resample('MS')
rs_obj
<pandas.core.resample.DatetimeIndexResampler object at 0x00000232EDE79730>
rs_obj.mean()
---------------------------------------------------------------------------
NotImplementedError Traceback (most recent call last)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1490, in GroupBy._cython_agg_general.<locals>.array_func(values)
1489 try:
-> 1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
1493 how,
1494 axis=data.ndim - 1,
1495 min_count=min_count,
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:959, in BaseGrouper._cython_operation(self, kind, values, how, axis, min_count, **kwargs)
958 ngroups = self.ngroups
--> 959 return cy_op.cython_operation(
960 values=values,
961 axis=axis,
962 min_count=min_count,
963 comp_ids=ids,
964 ngroups=ngroups,
965 **kwargs,
966 )
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:657, in WrappedCythonOp.cython_operation(self, values, axis, min_count, comp_ids, ngroups, **kwargs)
649 return self._ea_wrap_cython_operation(
650 values,
651 min_count=min_count,
(...)
654 **kwargs,
655 )
--> 657 return self._cython_op_ndim_compat(
658 values,
659 min_count=min_count,
660 ngroups=ngroups,
661 comp_ids=comp_ids,
662 mask=None,
663 **kwargs,
664 )
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:497, in WrappedCythonOp._cython_op_ndim_compat(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
495 return res.T
--> 497 return self._call_cython_op(
498 values,
499 min_count=min_count,
500 ngroups=ngroups,
501 comp_ids=comp_ids,
502 mask=mask,
503 result_mask=result_mask,
504 **kwargs,
505 )
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:541, in WrappedCythonOp._call_cython_op(self, values, min_count, ngroups, comp_ids, mask, result_mask, **kwargs)
540 out_shape = self._get_output_shape(ngroups, values)
--> 541 func = self._get_cython_function(self.kind, self.how, values.dtype, is_numeric)
542 values = self._get_cython_vals(values)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:173, in WrappedCythonOp._get_cython_function(cls, kind, how, dtype, is_numeric)
171 if "object" not in f.__signatures__:
172 # raise NotImplementedError here rather than TypeError later
--> 173 raise NotImplementedError(
174 f"function is not implemented for this dtype: "
175 f"[how->{how},dtype->{dtype_str}]"
176 )
177 return f
NotImplementedError: function is not implemented for this dtype: [how->mean,dtype->object]
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:1692, in _ensure_numeric(x)
1691 try:
-> 1692 x = float(x)
1693 except (TypeError, ValueError):
1694 # e.g. "1+1j" or "foo"
ValueError: could not convert string to float: 'CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC'
During handling of the above exception, another exception occurred:
ValueError Traceback (most recent call last)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:1696, in _ensure_numeric(x)
1695 try:
-> 1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
ValueError: complex() arg is a malformed string
The above exception was the direct cause of the following exception:
TypeError Traceback (most recent call last)
Cell In[51], line 1
----> 1 rs_obj.mean()
File ~\.conda\envs\JB\lib\site-packages\pandas\core\resample.py:979, in Resampler.mean(self, numeric_only, *args, **kwargs)
977 maybe_warn_args_and_kwargs(type(self), "mean", args, kwargs)
978 nv.validate_resampler_func("mean", args, kwargs)
--> 979 return self._downsample("mean", numeric_only=numeric_only)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\resample.py:1297, in DatetimeIndexResampler._downsample(self, how, **kwargs)
1293 return self.asfreq()
1295 # we are downsampling
1296 # we want to call the actual grouper method here
-> 1297 result = obj.groupby(self.grouper, axis=self.axis).aggregate(how, **kwargs)
1299 return self._wrap_result(result)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\generic.py:1269, in DataFrameGroupBy.aggregate(self, func, engine, engine_kwargs, *args, **kwargs)
1266 func = maybe_mangle_lambdas(func)
1268 op = GroupByApply(self, func, args, kwargs)
-> 1269 result = op.agg()
1270 if not is_dict_like(func) and result is not None:
1271 return result
File ~\.conda\envs\JB\lib\site-packages\pandas\core\apply.py:160, in Apply.agg(self)
157 kwargs = self.kwargs
159 if isinstance(arg, str):
--> 160 return self.apply_str()
162 if is_dict_like(arg):
163 return self.agg_dict_like()
File ~\.conda\envs\JB\lib\site-packages\pandas\core\apply.py:496, in Apply.apply_str(self)
494 if "axis" in arg_names:
495 self.kwargs["axis"] = self.axis
--> 496 return self._try_aggregate_string_function(obj, f, *self.args, **self.kwargs)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\apply.py:565, in Apply._try_aggregate_string_function(self, obj, arg, *args, **kwargs)
563 if f is not None:
564 if callable(f):
--> 565 return f(*args, **kwargs)
567 # people may try to aggregate on a non-callable attribute
568 # but don't let them think they can pass args to it
569 assert len(args) == 0
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1855, in GroupBy.mean(self, numeric_only, engine, engine_kwargs)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
-> 1855 result = self._cython_agg_general(
1856 "mean",
1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1507, in GroupBy._cython_agg_general(self, how, alt, numeric_only, min_count, **kwargs)
1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
-> 1507 new_mgr = data.grouped_reduce(array_func)
1508 res = self._wrap_agged_manager(new_mgr)
1509 out = self._wrap_aggregated_output(res)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\internals\managers.py:1503, in BlockManager.grouped_reduce(self, func)
1499 if blk.is_object:
1500 # split on object-dtype blocks bc some columns may raise
1501 # while others do not.
1502 for sb in blk._split():
-> 1503 applied = sb.apply(func)
1504 result_blocks = extend_blocks(applied, result_blocks)
1505 else:
File ~\.conda\envs\JB\lib\site-packages\pandas\core\internals\blocks.py:329, in Block.apply(self, func, **kwargs)
323 @final
324 def apply(self, func, **kwargs) -> list[Block]:
325 """
326 apply the function to my values; return a block if we are not
327 one
328 """
--> 329 result = func(self.values, **kwargs)
331 return self._split_op_result(result)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1503, in GroupBy._cython_agg_general.<locals>.array_func(values)
1490 result = self.grouper._cython_operation(
1491 "aggregate",
1492 values,
(...)
1496 **kwargs,
1497 )
1498 except NotImplementedError:
1499 # generally if we have numeric_only=False
1500 # and non-applicable functions
1501 # try to python agg
1502 # TODO: shouldn't min_count matter?
-> 1503 result = self._agg_py_fallback(values, ndim=data.ndim, alt=alt)
1505 return result
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1457, in GroupBy._agg_py_fallback(self, values, ndim, alt)
1452 ser = df.iloc[:, 0]
1454 # We do not get here with UDFs, so we know that our dtype
1455 # should always be preserved by the implemented aggregations
1456 # TODO: Is this exactly right; see WrappedCythonOp get_result_dtype?
-> 1457 res_values = self.grouper.agg_series(ser, alt, preserve_dtype=True)
1459 if isinstance(values, Categorical):
1460 # Because we only get here with known dtype-preserving
1461 # reductions, we cast back to Categorical.
1462 # TODO: if we ever get "rank" working, exclude it here.
1463 res_values = type(values)._from_sequence(res_values, dtype=values.dtype)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:994, in BaseGrouper.agg_series(self, obj, func, preserve_dtype)
987 if len(obj) > 0 and not isinstance(obj._values, np.ndarray):
988 # we can preserve a little bit more aggressively with EA dtype
989 # because maybe_cast_pointwise_result will do a try/except
990 # with _from_sequence. NB we are assuming here that _from_sequence
991 # is sufficiently strict that it casts appropriately.
992 preserve_dtype = True
--> 994 result = self._aggregate_series_pure_python(obj, func)
996 npvalues = lib.maybe_convert_objects(result, try_float=False)
997 if preserve_dtype:
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\ops.py:1015, in BaseGrouper._aggregate_series_pure_python(self, obj, func)
1012 splitter = self._get_splitter(obj, axis=0)
1014 for i, group in enumerate(splitter):
-> 1015 res = func(group)
1016 res = libreduction.extract_result(res)
1018 if not initialized:
1019 # We only do this validation on the first iteration
File ~\.conda\envs\JB\lib\site-packages\pandas\core\groupby\groupby.py:1857, in GroupBy.mean.<locals>.<lambda>(x)
1853 return self._numba_agg_general(sliding_mean, engine_kwargs)
1854 else:
1855 result = self._cython_agg_general(
1856 "mean",
-> 1857 alt=lambda x: Series(x).mean(numeric_only=numeric_only),
1858 numeric_only=numeric_only,
1859 )
1860 return result.__finalize__(self.obj, method="groupby")
File ~\.conda\envs\JB\lib\site-packages\pandas\core\generic.py:11556, in NDFrame._add_numeric_operations.<locals>.mean(self, axis, skipna, numeric_only, **kwargs)
11539 @doc(
11540 _num_doc,
11541 desc="Return the mean of the values over the requested axis.",
(...)
11554 **kwargs,
11555 ):
> 11556 return NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\generic.py:11201, in NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
11194 def mean(
11195 self,
11196 axis: Axis | None = 0,
(...)
11199 **kwargs,
11200 ) -> Series | float:
> 11201 return self._stat_function(
11202 "mean", nanops.nanmean, axis, skipna, numeric_only, **kwargs
11203 )
File ~\.conda\envs\JB\lib\site-packages\pandas\core\generic.py:11158, in NDFrame._stat_function(self, name, func, axis, skipna, numeric_only, **kwargs)
11154 nv.validate_stat_func((), kwargs, fname=name)
11156 validate_bool_kwarg(skipna, "skipna", none_allowed=False)
> 11158 return self._reduce(
11159 func, name=name, axis=axis, skipna=skipna, numeric_only=numeric_only
11160 )
File ~\.conda\envs\JB\lib\site-packages\pandas\core\series.py:4670, in Series._reduce(self, op, name, axis, skipna, numeric_only, filter_type, **kwds)
4665 raise TypeError(
4666 f"Series.{name} does not allow {kwd_name}={numeric_only} "
4667 "with non-numeric dtypes."
4668 )
4669 with np.errstate(all="ignore"):
-> 4670 return op(delegate, skipna=skipna, **kwds)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:96, in disallow.__call__.<locals>._f(*args, **kwargs)
94 try:
95 with np.errstate(invalid="ignore"):
---> 96 return f(*args, **kwargs)
97 except ValueError as e:
98 # we want to transform an object array
99 # ValueError message to the more typical TypeError
100 # e.g. this is normally a disallowed function on
101 # object arrays that contain strings
102 if is_object_dtype(args[0]):
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:158, in bottleneck_switch.__call__.<locals>.f(values, axis, skipna, **kwds)
156 result = alt(values, axis=axis, skipna=skipna, **kwds)
157 else:
--> 158 result = alt(values, axis=axis, skipna=skipna, **kwds)
160 return result
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:421, in _datetimelike_compat.<locals>.new_func(values, axis, skipna, mask, **kwargs)
418 if datetimelike and mask is None:
419 mask = isna(values)
--> 421 result = func(values, axis=axis, skipna=skipna, mask=mask, **kwargs)
423 if datetimelike:
424 result = _wrap_results(result, orig_values.dtype, fill_value=iNaT)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:727, in nanmean(values, axis, skipna, mask)
724 dtype_count = dtype
726 count = _get_counts(values.shape, mask, axis, dtype=dtype_count)
--> 727 the_sum = _ensure_numeric(values.sum(axis, dtype=dtype_sum))
729 if axis is not None and getattr(the_sum, "ndim", False):
730 count = cast(np.ndarray, count)
File ~\.conda\envs\JB\lib\site-packages\pandas\core\nanops.py:1699, in _ensure_numeric(x)
1696 x = complex(x)
1697 except ValueError as err:
1698 # e.g. "foo"
-> 1699 raise TypeError(f"Could not convert {x} to numeric") from err
1700 return x
TypeError: Could not convert CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC to numeric
# We can chain all of that together
df_mm = df.resample('MS').mean()
df_mm[['T_DAILY_MIN', 'T_DAILY_MEAN', 'T_DAILY_MAX']].plot()
And this concludes this notebook’s tutorial 😀
If you would like to learn more about pandas, check out the groupby tutorial from the Earth and Environmental Data Science book, the Github repository of the Python for Data Analysis, or the pandas community tutorials.
